Testing for RET is essential to identify patients who may be eligible for Retevmo1

NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) recommend testing for RET alterations in appropriate patients with advanced and/or metastatic NSCLC and thyroid carcinoma* to determine if they are eligible for RET inhibitors such as selpercatinib (Retevmo)2,3
NCCN makes no warranties of any kind whatsoever regarding their content, use or application and disclaims any responsibility for their application or use in any way.
Next-generation sequencing (NGS) can be an accurate and tissue-efficient method to test for driver RET alterations and other targetable biomarkers4-7†
Retevmo may affect both healthy cells and tumor cells, which can result in side effects, some of which can be serious.1
- Both RET point mutations and fusions can be detected by NGS.4-6
- Immunohistochemistry (IHC) is not preferred for detecting RET alterations due to low sensitivity and variable specificity9,10
- Test the tissue: molecular testing of FFPE tumor tissue specimens is preferred for detecting RET fusions and point mutations8,11-13

- In the clinical trial, identification of a RET gene alteration was prospectively determined in local laboratories using NGS, PCR, or FISH1
- IHC testing was not used in LIBRETTO-0011
Why NGS?
Broad molecular profiling to identify appropriate targeted therapies can improve outcomes in NSCLC14
- NCCN Guidelines for NSCLC recommend that, when feasible, molecular testing of NSCLC specimens be performed via a broad, panel-based approach, most typically performed by NGS2
- Because of potential tissue limitations in metastatic NSCLC and the increased number of actionable biomarkers, NGS testing is part of the most comprehensive strategy to identify appropriate targeted therapies7
- Consider NGS testing to identify the 69% of patients with lung adenocarcinoma who have a potentially actionable oncogenic driver alteration and may benefit from appropriate approved or investigational targeted therapy15,16
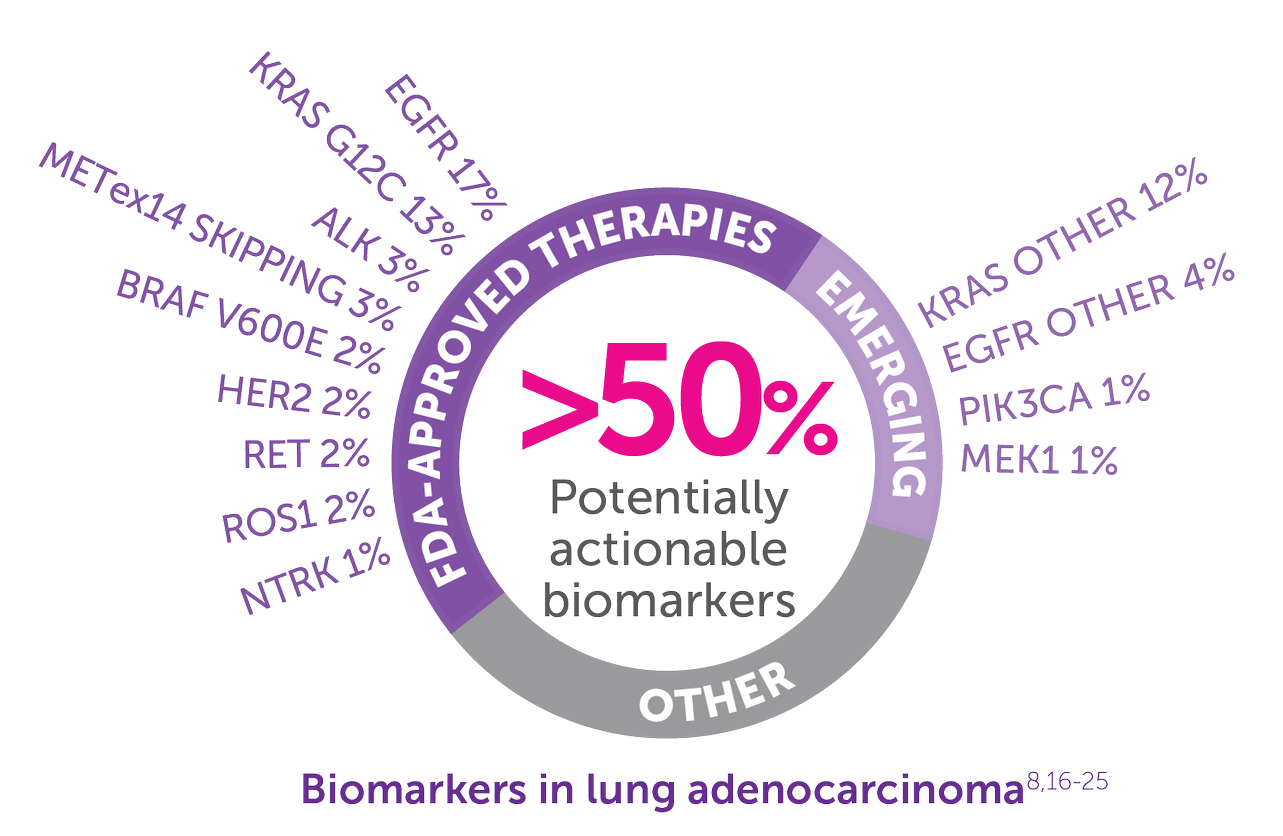
Emerging=biomarkers with therapies under investigation but not approved.
Other=unknown oncogenic driver detected.16
EGFR=EGFR sensitizing mutations including exon 20 insertions.16,23
EGFR other=secondary EGFR mutations, including Thr790Met and Cys797Ser, and other less common EGFR mutations.16
KRAS other=all KRAS mutations other than KRAS G12C.16,22
NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®)
For NSCLC:
- recommend testing for RET fusions in eligible patients with metastatic non-small cell lung cancer2
- recommend molecular testing and strongly advise broad molecular profiling for multiple biomarkers, including RET, in eligible patients with metastatic NSCLC2*‡
*The NCCN Guidelines provide recommendations for certain individual biomarkers that should be tested and recommend testing techniques but do not endorse any specific commercially available biomarker assays or commercial laboratories.
†Through design and validation, the test has established high sensitivity, specificity, and reproducibility for the detection of genomic alterations.
‡It is recommended at this time that, when feasible, testing be performed via a broad, panel-based approach, most typically performed by NGS. For patients who, in broad panel testing, don’t have identifiable driver oncogenes (especially in never smokers), consider RNA-based NGS, if not already performed, to maximize detection of fusion events.
For Thyroid Carcinoma:
- recommend molecular testing for RET fusions and RET point mutations for certain patients with advanced or metastatic thyroid carcinomas3
Consider waiting for RET test results before making therapeutic decisions1
NGS testing
- NGS testing for RET fusions: when properly designed, NGS testing is able to detect known and unknown fusion events6
- A combination of RNA- and DNA-based NGS testing may be a more comprehensive approach to identify oncogenic drivers missed by DNA-based NGS alone24
Most common RET fusion partners identified in the LIBRETTO-001 phase I/II clinical trial:
NSCLC25:
- 59% KIF5B
- 22% CCDC6
- 11% Unknown§
- 6% Otherǁ
- 2% NCOA4
Thyroid cancer other than MTC26:
- 52% CCDC6
- 33% NCOA4
- 15% Other¶
§Unknown includes positive by FISH or PCR.
ǁOthers included KIAA1468(2), ARHGAP12, CCDC88C, CLIP1, DOCK1+RBPMS, ERC1, PRKAR1A, and TRIM24 (all 1 each).25
¶Others included CCDC1686, ERC1, KTN1, and RUFY (all 1 each).26
NGS testing
- NGS allows for multiplex testing on a small amount of tissue for the detection of rare, as well as common, cancer-related biomarkers4,7,27
- RET point mutations can be detected by NGS4-6
Most common RET mutations identified in the LIBRETTO-001 phase I/II clinical trial26:
- 57% M918T
- 19% Extracellular cysteine mutations
- 16% Other#
- 8% V804M/L
#Others included D631-L633delinsE(5), E632-L633del(4), A883F(4), D631-L633delinsV(2), L790F(2), D898-E901del(2), D898_E901del + D903_S904delinsEP, K666N, T636-V637insCRT, D378-G385delinsE (all 1 each).26

- While not recommended as a replacement for a diagnostic tissue biopsy, consider liquid biopsy when FFPE tumor tissue is unavailable or insufficient for molecular profiling11
- While a positive liquid biopsy result is considered reliable, a negative result requires confirmation with tumor tissue testing11
-
NCCN Guidelines: principles of molecular and biomarker analysis in metastatic NSCLC2
- Plasma ctDNA (liquid biopsy) should not be used in lieu of a histologic tissue diagnosis
- Studies have demonstrated liquid biopsy to generally have very high specificity but significantly compromised sensitivity, with a false-negative rate of up to 30%
- Standards and guidelines for liquid biopsy testing for genetic alterations have not been established
Test for RET1
Ensure your test can detect driver RET fusions in NSCLC and non-medullary thyroid cancer and driver RET mutations in MTC
ALK=anaplastic lymphoma kinase; BRAF=v-raf murine sarcoma viral oncogene homolog B; DNA=deoxyribonucleic acid; EGFR=epidermal growth factor receptor; FFPE=formalin-fixed paraffin-embedded; HER2=human epidermal growth factor receptor 2; KRAS=Kirsten rat sarcoma; MEK1=dual specificity mitogen-activated protein kinase kinase 1; METex14=mesenchymal-epithelial transition exon 14 skipping; MTC=medullary thyroid cancer; NCCN=National Comprehensive Cancer Network; NSCLC=non-small cell lung cancer; NTRK=neurotrophic receptor tyrosine kinase; PIK3CA=phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha; PTC=papillary thyroid cancer; RET=rearranged during transfection; RNA=ribonucleic acid; ROS1=reactive oxygen species 1.
References: 1. Retevmo (selpercatinib). Prescribing Information. Lilly USA, LLC. 2. Referenced with permission from The NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Non-Small Cell Lung Cancer V5.2022. © National Comprehensive Cancer Network, Inc. 2022. All rights reserved. Accessed December 1, 2022. To view the most recent and complete version of the guidelines, go online to https://www.nccn.org. 3. Referenced with permission from The NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Thyroid Carcinoma V3.2022. © National Comprehensive Cancer Network, Inc. 2022. All rights reserved. Accessed December 1, 2022. To view the most recent and complete version of the guidelines, go online to https://www.nccn.org. 4. Gregg JP, Li T, Yoneda KY. Molecular testing strategies in non-small cell lung cancer: optimizing the diagnostic journey. Transl Lung Cancer Res. 2019;8(3):286-301. 5. Suh JH, Schrock AB, Johnson A, et al. Hybrid capture-based comprehensive genomic profiling identifies lung cancer patients with well-characterized sensitizing epidermal growth factor receptor point mutations that were not detected by standard of care testing. Oncologist. 2018;23(7):776-781. 6. Mertens F, Johansson B, Fioretos T, et al. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15(6):371-381. 7. Suh JH, Johnson A, Albacker L, et al. Comprehensive genomic profiling facilitates implementation of the National Comprehensive Cancer Network Guidelines for lung cancer biomarker testing and identifies patients who may benefit from enrollment in mechanism-driven clinical trials. Oncologist. 2016;21(6):684-691. 8. Drilon A, Hu ZI, Lai GGY, et al. Targeting RET-driven cancers: lessons from evolving preclinical and clinical landscapes. Nat Rev Clin Oncol. 2018;15(3):151-167. 9. Ferrara R, Auger N, Auclin E, et al. Clinical and translational implications of RET rearrangements in non-small cell lung cancer. J Thorac Oncol. 2018;13(1):27-45. 10. Naidoo J, Drilon A. Molecular diagnostic testing in non-small cell lung cancer. Am J Hematol Oncol. 2014;10(4):4-11. 11. Rolfo C, Mack PC, Scagliotti GV, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol. 2018;13(9):1248-1268. 12. Dietel M, Bubendorf L, Dingemans A-M, et al. Diagnostic procedures for non-small-cell lung cancer (NSCLC): recommendations of the European Expert Group. Thorax. 2016;71(2):177-184. 13. Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol. 2018;13(3):323-358. 14. Singal G, Miller PG, Agarwala V, et al. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a clinicogenomic database. JAMA. 2019;321:1391-1399. 15. Fernandes MGO, Jacob M, Martins N, et al. Targeted gene next-generation sequencing panel in patients with advanced lung adenocarcinoma: paving the way for clinical implementation. Cancers (Basel). 2019;11(9):1229. 16. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299-311. 17. Drilon A, Oxnard GR, Tan DSW, et al. Efficacy of selpercatinib in RET fusion–positive non–small-cell lung cancer. N Engl J Med. 2020;383(9):813-824. 18. Amatu A, Sartore-Bianchi A, Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open. 2016;1(2):e000023. doi:10.1136/esmoopen-2015-000023. 19. Vansteenkiste JF, Van De Kerkhove C, Wauters E, et al. Capmatinib for the treatment of non-small cell lung cancer. Expert Rev Anticancer Ther. 2019;19(8):659-671. 20. FDA approves first targeted therapy to treat aggressive form of lung cancer. News release. FDA. May 6, 2020. Accessed May 21, 2020. https://www.fda.gov/news-events/press-announcements/fda-approves-first-targeted-therapy-treat-aggressive-form-lung-cancer. 21. Planchard D, Besse B, Groen HJM, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol. 2016;17(7):984-993. 22. FDA approves first targeted therapy for lung cancer mutation previously considered resistant to drug therapy. News release. FDA. May 28, 2021. Accessed June 17, 2021. https://www.fda.gov/news-events/press-announcements/fda-approves-first-targeted-therapy-lung-cancer-mutation-previously-considered-resistant-drug. 23. FDA approves first targeted therapy for subset of non-small cell lung cancer. News release. FDA. May 21, 2021. Accessed June 30, 2021. https://www.fda.gov/news-events/press-announcements/fda-approves-first-targeted-therapy-subset-non-small-cell-lung-cancer. 24. Chevallier M, Borgeaud M, Addeo A, Friedlaender A. Oncogenic driver mutations in non-small cell lung cancer: past, present and future. World J Clin Oncol. 2021;12(4):217-237. 25. Zhao Z, Yang, X. Targeting HER2 alterations in non–small-cell lung cancer: a comprehensive review. JCO Precis Oncol. 2020;4:411-425. doi:10.1200/PO.19.00333. 26. Benayed R, Offin M, Mullaney K, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res. 2019;25(15):4712-4722. 27. Wells SA Jr, Asa SL, Dralle H, et al. Revised American Thyroid Association guidelines for the management of medullary thyroid carcinoma. Thyroid. 2015;25(6):567-610. 28. Agrawal N, Jiao Y, Sausen M, et al. Exomic sequencing of medullary thyroid cancer reveals dominant and mutually exclusive oncogenic mutations in RET and RAS. J Clin Endocrinol Metab. 2013;98(2):E364-E369. 29. Simbolo M, Mian C, Barollo S, et al. High-throughput mutation profiling improves diagnostic stratification of sporadic medullary thyroid carcinomas. Virchows Arch. 2014;465(1):73-78.
INDICATIONS
Retevmo is a kinase inhibitor indicated for the treatment of:
- adult patients with locally advanced or metastatic non-small cell lung cancer (NSCLC) with a rearranged during transfection (RET) gene fusion, as detected by an FDA-approved test
- adult and pediatric patients 12 years of age and older with advanced or metastatic medullary thyroid cancer (MTC) with a RET mutation, as detected by an FDA-approved test, who require systemic therapy*
- adult and pediatric patients 12 years of age and older with advanced or metastatic thyroid cancer with a RET gene fusion, as detected by an FDA-approved test, who require systemic therapy and who are radioactive iodine-refractory (if radioactive iodine is appropriate)*
- adult patients with locally advanced or metastatic solid tumors with a RET gene fusion that have progressed on or following prior systemic treatment or who have no satisfactory alternative treatment options*
*These indications are approved under accelerated approval based on overall response rate (ORR) and duration of response (DoR). Continued approval for these indications may be contingent upon verification and description of clinical benefit in confirmatory trials.